Question 1: Which statement about donor selection for allogeneic hematopoietic cell transplant is correct?
Correct Answer: C. If a donor is available, the genotypic identity between the patient and his brother should be established by typing the patient’s parents; if this is not possible, the brother should be HLA-matched at a minimum for HLA-A, HLA-B, HLA-C, and HLA-DRB1 at high resolution (second field).
Expert Perspective
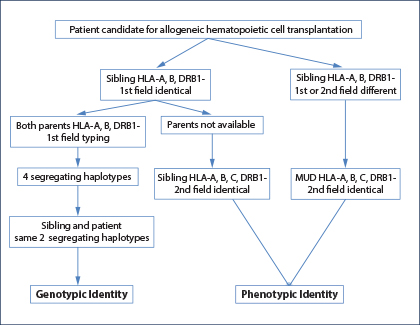
Fig. 1: HLA-typing strategies to identify an HLA genotypically identical sibling, an HLA phenotypically identical sibling, or an HLA-matched unrelated donor.
An HLA-identical or “fully” HLA-matched sibling donor is considered the optimal and first choice graft for allogeneic hematopoietic cell transplant.1-3 The genotypic identity between the two siblings can only be established by typing both parents to understand segregation of the four haplotypes (Fig. 1). In the absence of family studies, even if the patient and his brother are identical for HLA-A, HLA-B, and HLA-DRB1 at low resolution (first field), it is possible they are encoded by different haplotypes and therefore “hide” allelic mismatches. If genotypic identity cannot be established, the patient and his brother should be considered no different from two unrelated individuals, and therefore a minimum matching at high resolution (second field) for HLA-A, HLA-B, HLA-C, and HLA-DRB1 should be established (choice C; Fig. 1).
Question 2: Which statement about the concept of linkage disequilibrium within the HLA system is correct?
Correct Answer: C. Linkage disequilibrium implies that certain alleles occur together with a higher frequency than expected by chance.
Expert Perspective
Linkage disequilibrium implies that certain alleles occur together with a greater frequency than expected by chance (nonrandom gametic association4; choice C). In general, linkage disequilibrium is more frequently observed between loci that are in close proximity. Linkage disequilibrium is a hallmark of both HLA class I (in particular HLA-B and HLA-C loci) and HLA class II (in particular HLA-DR and DQ loci). Although there is strong linkage disequilibrium between the DPA1 and the DPB1 loci, due to a recombination hotspot telomeric to HLA-DP, the linkage disequilibrium between HLA-DP and the other HLA class II loci is rather low. For this reason, matching for HLA-DR and DQ in two unrelated individuals is frequently not accompanied by matching for HLA-DP.

Syed A. Abutalib, MD
On the other hand, linkage disequilibrium is at the basis of the probability for an individual to identify a HLA-matched unrelated donor: In patients with frequent haplotypes such as HLA-A1, HLA-B8, and HLA-DR3, the probability is much higher than average, whereas in patients with unusual linkage disequilibrium, for instance through recombination, the probability drops dramatically, and clinicians may be advised to immediately consider a mismatched unrelated or family donor.5 In certain cases, linkage disequilibrium may also help to predict allelic resolution; however, this is not completely accurate, particularly in ethnic groups whose HLA types have not yet been well studied. For instance, more than 95% of white individuals who carry HLA-B*07:02 will have HLA-C*07:02, whereas those who carry B*18:01 may have either C*07:01, C*12:03, or C*05:01.
Question 3: Which of the following unrelated donors would be the first choice for allogeneic hematopoietic cell transplantation?
Correct Answer: C. HLA-A, HLA-B, HLA-C, and HLA-DRB1 matched at the allele level with the recipient.

Katharina Fleischhauer, MD
Expert Perspective
Approximately 70% of patients in need of allogeneic hematopoietic cell transplant will not have an HLA-identical sibling. Presently, an HLA-matched unrelated donor is considered the preferred donor for these patients. Based on the findings of numerous large, contemporary retrospective studies and comprehensive reviews, overall survival is increased and transplantation-related mortality reduced when recipients of unrelated donor allogeneic hematopoietic cell transplant are matched at high resolution (second field) for HLA-A, HLA-B, HLA-C, and HLA-DRB1 at the allele level (eight of eight HLA-matched; choices A, B, and C).1,3,6 The likelihood of finding an HLA-matched unrelated donor ranges from 75% to 90% for patients of European descent down to 16% for blacks of South or Central American descent.2,7
Question 4: Which statement about high-resolution (second field) HLA typing is correct?
Answer: C. High-resolution HLA typing defines sets of alleles that encode the same protein sequence for the HLA molecule’s antigen-binding site.
Expert Perspective

Fig. 2: HLA nomenclature for full alleles (upper panel), P Groups or G Groups (lower left and right panel, respectively). This figure was kindly provided by Professor Steven GE Marsh, of the Anthony Nolan Research Institute, London UK (www.hla.alleles.org).9 Low- and high-resolution typing refer to the first and second fields, respectively. Serologic typing corresponds generally to the first field, although some exceptions apply to certain serologic splits, which can be defined molecularly only at the second-field level. For selection of a phenotypically HLA-identical donor (see Figure 1), typing to the second field with ambiguity resolution of the P or G groups is sufficient.
The adoption of high-resolution (second field) HLA allele–level typing for screening unrelated donors at enrollment has been associated with quicker identification of suitable donors, resulting in a tangible clinical benefit for patients.8 High-resolution typing defines sets of alleles that encode the same protein sequence for the HLA molecule’s antigen-binding site (eg, HLA-A*02:01P; choice C). In contrast, low-resolution (first field) typing provides information regarding the allele group but not the specific HLA protein (eg, HLA-A*029; Fig. 2; choice B).
High-resolution typing is generally performed by direct sequencing, either by classical Sanger methods or more recently by next-generation sequencing.10 The latter method reduces, but does not always eliminate, typing ambiguities arising from the same combination of polymorphisms occurring in different sets of heterozygous HLA alleles in cis and trans positions. Typing ambiguities should be resolved at least for those alleles reported as common or well documented.11 High-resolution HLA typing can be performed on a peripheral blood specimen (choice A), although DNA from other tissues including saliva is also suitable.
For patients with active leukemia who present for HLA typing, the possibility of genomic loss of heterozygosity by leukemic blasts as a mode of evasion from the host’s immune system should be considered, particularly in the case of homozygosity.12,13 It is therefore mandatory for clinicians to indicate to the laboratory whether or not the blood sent for HLA typing was obtained from the patient in remission and to repeat typing on a sample in remission especially in the case of homozygosity. Saliva or buccal swaps can be used, but it should be noted that infiltration of the gingiva by leukemic blasts has been described. It is always mandatory to confirm the patient’s and donor’s HLA type at least for the loci HLA-A, HLA-B, HLA-C, and HLA-DRB1 once on a new sample prior to transplantation.
Question 5: Which statement would be correct if multiple HLA-matched (eight of eight) unrelated donors were available?
Answer: B. HLA-DPB1 matching status would be considered to identify permissive mismatches.
Expert Perspective
When multiple unrelated donors matched at high resolution (second field) for HLA-A, HLA-B, HLA-C, and HLA-DRB1 (eight of eight) are available, the consideration of additional HLA loci and/or non-HLA factors may be used to prioritize the final donor selection (choice A).1-3 Testing of additional HLA loci (eg, HLA-DPB1, HLA-DQB1, and HLA-DRB3/4/5) will assist in selecting donors with minimal mismatching at low expression loci14 and for HLA-DPB1, low expression variants15 or T-cell epitope permissive HLA-DPB1 mismatches.16-19 The latter algorithm is freely available as a T-cell epitope webtool at the IMGT (International Immunogenetics Information System) database (www.ebi.ac.uk/ipd/imgt/hla/dpb.html) and has recently been included in the donor-search algorithms of the Be The Match Registry20 as well as the German Registry.21
Testing for antibodies specific to donor HLA (donor-specific antibodies) can also support the selection of appropriate donors in the context of HLA-sensitized patients to avoid the potential risk of graft failure.22,23 In seven of eight HLA-matched settings, donors can also be prioritized for permissive HLA-C*03:03/03:04 mismatches24 or host-vs-graft–only mismatches25 to optimize outcomes.
Finally, based upon the findings of a recently reported retrospective analysis on behalf of the Center for International Blood and Marrow Transplant Research,26 centers may consider the following: (1) younger HLA-matched unrelated donors, found to be associated with less acute graft-vs-host disease and better overall survival; (2) ABO-compatible donors, which were also associated with a modest survival benefit; and (3) cytomegalovirus-serostatus–matched donors.27 ■
Disclosure: Dr. Abutalib reported no potential conflicts of interest. Dr. Fleischhauer reported this work was supported by grants from the Deutsche José Carreras Leukämie Stiftung, the European Commission Transcan JTC2012, and Joseph Senker Stiftung.
References
9. Marsh SG, et al: An update to HLA nomenclature, 2010. Bone Marrow Transplant 45:846-848, 2010.
10. Gabriel C, et al: HLA typing by next-generation sequencing. Tissue Antigens 83:65-75, 2014.
11. Mack SJ, et al: Common and well-documented HLA alleles. Tissue Antigens 81:194-203, 2013.

